Co-developed at the Italian National Council of Research Institute of Clinical Physiology (IFC-CNR) and the Research Center "E. Piaggio", University of Pisa, the freeware can be downloaded at this link. The source code can be downloaded at this link.
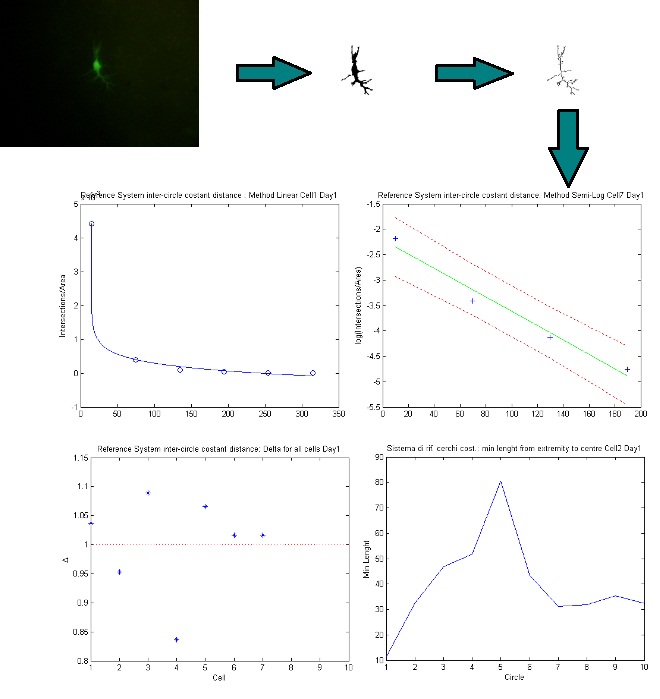
NEMO (NEuron MOrphological analysis tool) is a new freeware for semi automated quantitative and dynamic analysis of neuron morphometry. It incorporates the most important microstructural quantification methods, such as fractal and sholl analysis with statistical and classification tools to provide an integrated image processing environment which enables fast and easy feature identification.
It includes:
- Friendly interactive graphical user interface
- Image pre-processing
- Morphological analysis
- Topological analysis
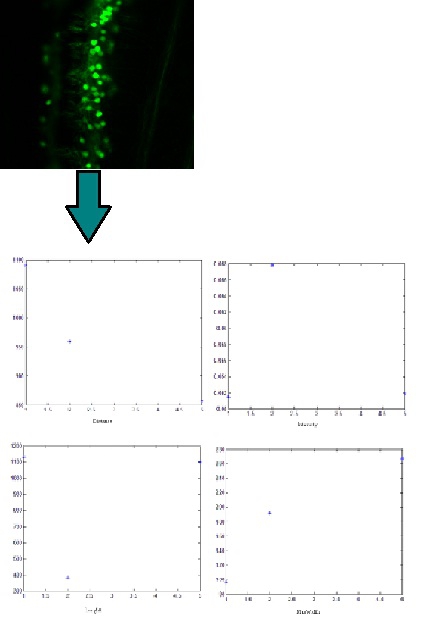
- Cell counting
- *3-way PCA analysis (also available as an ImageJ plugin)
- Plot of variables
Sequential images of labeled or unlabelled neurons or tissue slices can be uploaded batch-wise in order to create a 3 axis (time, image coordinate) data base and a datamatrix of variables for 3-way Principal Component Analysis*.
For additional information contact: chiara.magliaro@googlemail.com or lucia.billeci@ifc.cnr.it
Authors: Billeci Lucia, Magliaro Chiara, Giovanni Pioggia, Arti Ahluwalia Copyrighted 13/12/2011, SIAE # 008241/D007440
References
- Billeci L., Magliaro C.,Pioggia G., Ahluwalia A., "NEuronMOrphological analysis tool: open-source software for quantitative morphometrics", Frontiers in NeuroInformatics, 2013.
Users are kindly asked to cite the paper and the software source.
- Billeci L., Pioggia G., Vaglini F., Ahluwalia A., "Automated extraction and classification of dynamic metrical features of morphological development in dissociated Purkinje neurons", J Neurosci Methods. 2010:185(2):315-24
- Billeci L., Pioggia G., Vaglini F., Ahluwalia A., "Assessment and comparison of neural morphology through metrical feature extraction and analysis in neuron and neuron-glia cultures", J Biol Phys. 2009, 35(4):447-64

